Outline
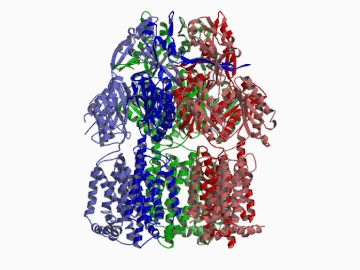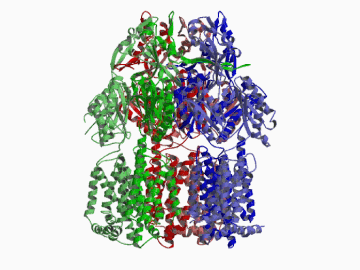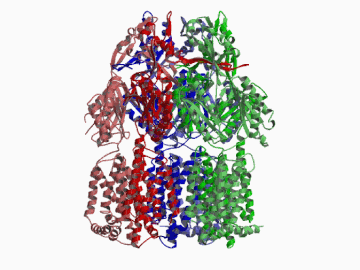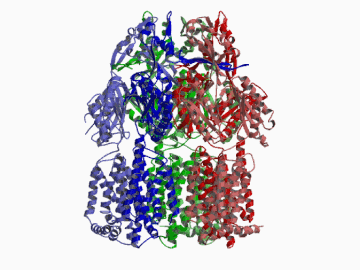
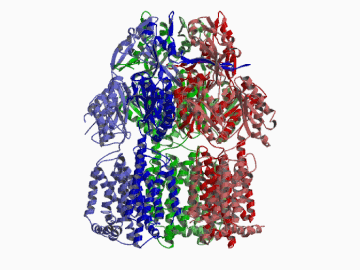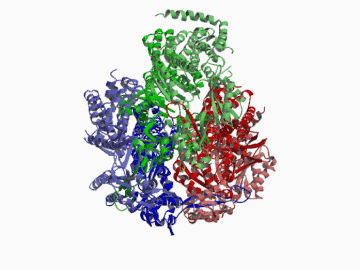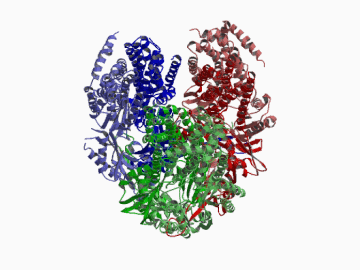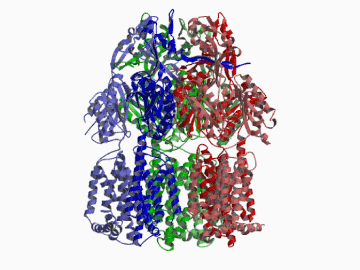
Xenobiotic extruding pumps have recently been known to be widely distributed in living organisms from mammalian to bacteria as a host-defence mechanism in cellular level. These pumps not only confer multidrug resistance of cancer cells and pathogenic bacteria but also cause hereditary diseases through the mutation. The purposes of our laboratory is to elucidate the molecular structures and the molecular mechanisms of these xenobiotic exporters and the roles of these exporters in cell functions. In addition, the exporters having xenobiotic exporter-like molecular structures are identified in brain and platelets in order to elucidate the possible roles of exporters in intercellular signal transduction. In 2002, we successfully reported the article in Nature of the high resolution crystal structure of the xenobiotic exporter AcrB, that is a trimer having jellyfish like shape. The structure well explains TolC-docking, dual entrance model and possible proton penetrating pathways. This is the first case for the crystal structure determination of multidrug transporters as well as proton coupling one and gives a structural basis for understanding the membrane transport mechanism.
Current Research Programs
1. Crystallization and crystallographic analysis of xenobiotic exporters.
We succeeded in determining the first crystal structure of bacterial multidrug efflux transporter AcrB in 2002 and published in Nature 407, 971-977 (2002). This is not only the first crystal structure of a multidrug exporter but also the first atomic level structure of a proton-coupled secondary active transporter. It was a real milestone for understanding drug-microbe interactions and secondary active transport mechanism. We are now making effort to solve the structure of co-crystals of AcrB and its substrates.


2. Protein engineering analysis of the structure and function of xenobiotic exporters
We are performing the site-directed mutagenesis studies based on the crystal structure of the xenobiotic exporter AcrB. AcrB trimer has a central pore in the periplasmic head piece. Cysteine-scanning mutagenesis studies on the pore helix revealed the importance of the inner wall of the pore for the substrate translocation. In addition, the site-directed mutagenesis of the conserved charged residues in AcrB revealed that the essential charged residues form ion pairs in the transmembrane region probably making proton pathway. We are now performing mutagenesis studies on the other structurally-interesting regions such as TolC-docking domain, AcrA-binding cleft, substrate-binding phenylalanine cluster, vestibules into the central cavity, and transmembrane groove. We hope that these studies help understanding how the structure acts as a drug exporter.
3. Systematic cloning of whole E. coli putative exporter genes and the genes of two-component signal transduction systems
We constructed the set of E. coli 37 putative drug exporter gene library. Among them, 20 genes actually encode drug exporters. We are studying these exporters one by one in detail. In addition, we first found that the bacterial two-component signal transduction systems control the expression of drug exporter genes and confer multidrug resistance. This is a completely new types of mechanism acquiring multidrug resistance.
4. Identification of novel exporter genes in brain and platelets and the studies on the physiological roles of these transporters using cultured cells and knockout mice
We constructed the knockout mice of a novel ABCA-type gene, which is mainly expressed in brain and testis. The knockout mice die at about 11 weeks after birth by myocardiopathy. We are now studying the cause of the phenotype and the biochemical characterization of the ABCA gene. In addition, we found that sphingosine 1-phosphate is exported from platelet probably by ABC-type transporters. We are now making effort to identify the transporter.